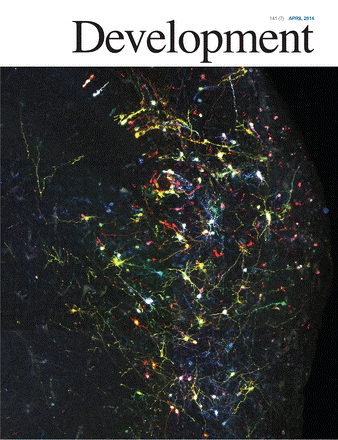
There are several methods available to label the entire progeny of a given stem or progenitor cell, but all of them have some drawbacks and could suffer from either underestimating or overestimating the actual size of the clones. To minimize such inaccuracies, most existing methods should be used with very few clones in a given specimen, impeding the investigations on generation, integration and relationship of multiple clones.
The groups of García-Moreno, Begbie and Molnár (all from DPAG) have just published a paper in the journal Development reporting CLoNe, a new method for clonal cell labelling in vivo. This method is based on the combined labelling (electroporation) of progenitor cells with three types of plasmids encoding (a) multiple fluorescent reporters, (b) a Cre driver and (c) a transposase. The method is developed so it allows the reliable tracing to specific progenitor populations, and the approach was tested in the mouse and chick embryonic telencephalon and in the chick limb bud. It is shown to permit long-term tracing through somatic integration of the reporter plasmids into the host cell genome, and to generate an unbiased colour distribution to maximize clone assignment.
CLoNe allows comparative analysis of progenitor cells across species and it is suitable for various tissues, as evidenced in muscular and epithelial tissue, and systems across several vertebrate species.
García-Moreno F, Vasistha NA, Begbie J, Molnár Z.
CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. (2014) Development. 141(7):1589-98. doi: 10.1242/dev.105254.
http://dev.biologists.org/content/141/7/1589.abstract
http://dev.biologists.org/content/141/7.cover-expansion
Supplementary reading
http://thenode.biologists.com/studying-genealogy-in-cell-clones/research/